Advantages
- Membrane‑context readout: Localization of protein interactions at the cell membrane offers a more physiologically relevant measurements of protein-protein interactions (PPI)
- Simple, modular anchors: Gap1C (native FWC motif) and Alp1C±FWC tails deliver consistent membrane docking and can be mixed/matched across split‑FP pairs.
- Works with flow cytometry (quantitative) and confocal microscopy (spatial localization)
- Screening‑ready: Demonstrated FACS‑based selection of dim/bright GFP1‑10 variants directly in the membrane‑proximal context.
Summary
Protein-protein interactions involving membrane proteins are notoriously difficult to study under physiologically relevant conditions. Many established methods focus on nuclear or cytosolic interactions and cannot fully replicate the native membrane context. Conventional split fluorescent protein systems often suffer from inefficiencies in reconstitution, potential over-expression artifacts, and challenges in targeting specific membrane regions. Additionally, extraneous background signals or nonspecific binding can cloud the interpretation of results, especially when attempting to track transient or weak interactions. As such, researchers require strategies that effectively anchor proteins at the membrane without compromising their natural conformations or interaction domains to ensure that the observed interactions reflect the true cellular environment.
Our inventors developed a new technology for detecting intracellular protein interactions by anchoring split‑fluorescent reporters to the inner surface of the yeast plasma membrane, enabling fluorescence only when two proteins come together. In this system, the split reporters (e.g., GFP1‑10/GFP11 or Cerulean1‑10/GFP11) are fused to membrane‑tethering tails—Gap1C (which natively includes an FWC palmitoylation motif) or Alp1C (optionally upgraded to Alp1C+FWC)—and either fragment can be paired with any of these anchors to co‑localize at the membrane and reconstitute signal.
Validation across multiple designs showed clear membrane rings by confocal imaging and strong flow‑cytometry reconstitution, with the highest signals when both fragments were tethered or when Alp1C carried the FWC motif; even weaker reporters like split Cerulean produced visible membrane localization. The platform also enables FACS‑based selection of bright and dim GFP1‑10 variants directly in this membrane‑proximal context, supporting screening and optimization workflows.
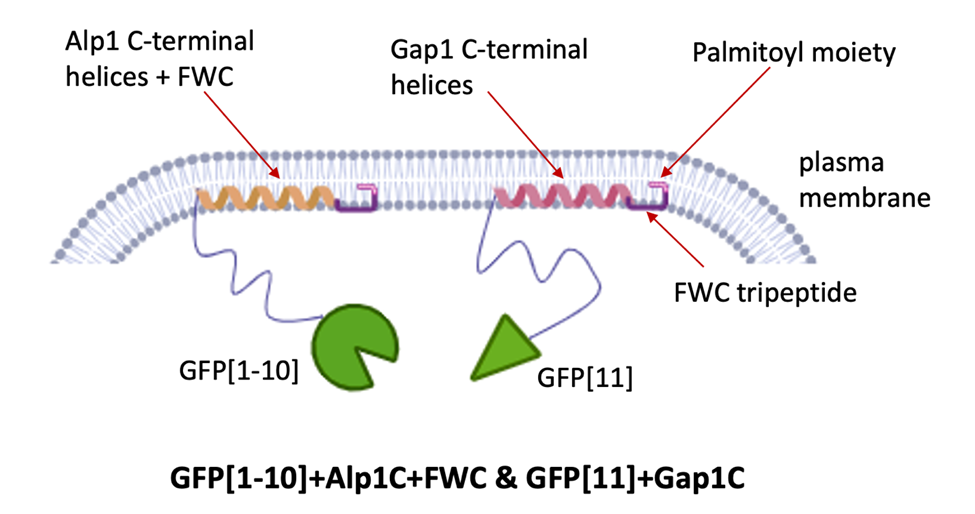
Schematic for Alp1C+FWC and Gap1C -terminal domains docking to the plasma membrane.
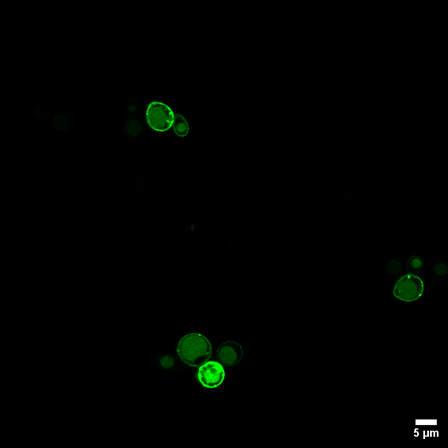
Membrane‑Localized Split‑FP Reconstitution in Yeast. Confocal microscopy shows strong GFP fluorescence forming a clear ring at the plasma membrane when both split‑GFP fragments are anchored using Gap1C, demonstrating robust intracellular co‑localization and reconstitution.
Desired Partnerships
- License
- Sponsored Research
- Co-Development