Advantages
- IPF Biomarker assay: 6-gene transcriptomic signature strongly correlates with disease severity and mortality across multiple patient cohorts and sample types
- Non-Invasive Testing: PCR test requires only a blood sample
- Potential for Precision-Based Therapies: gene-signature panel opens avenues to test targeted therapeutic interventions
- Identification of Drug Candidates: Identified several drug categories that can potentially reverse the harmful gene expression
Summary
Idiopathic Pulmonary Fibrosis (IPF) is a chronic, progressive lung disease characterized by scarring of lung tissue, leading to severe respiratory dysfunction and high mortality rates. Traditional biomarkers and prognostic indicators have limited ability to predict disease progression or guide personalized treatment.
Our researchers discovered a specific immune cell subtype, CD14+CD163-HLA-DRlow monocytes, with a 230-gene transcriptomic signature that is strongly associated with IPF severity and mortality. High-risk patients displayed decreased expression of T-cell co-stimulatory genes and higher expression of profibrotic, proangiogenic, and chemotactic factors. This signature was validated across multiple independent cohorts and biological samples, including peripheral blood, bronchoalveolar lavage (BAL), and lung tissue. A subset 6-gene version of the signature was developed, which can be measured through a simple blood test, making it suitable for clinical biomarker assay development such as an easy PCR blood test.
Using this gene signature, the researchers also identified drug categories capable of reversing the high-risk gene expression pattern, offering a new method to conduct drug discovery and identifying novel therapeutic targets for IPF. This provides a promising direction for developing precision-based therapies aimed at modulating the specific monocyte subtype and its associated genes to improve prognosis in IPF.
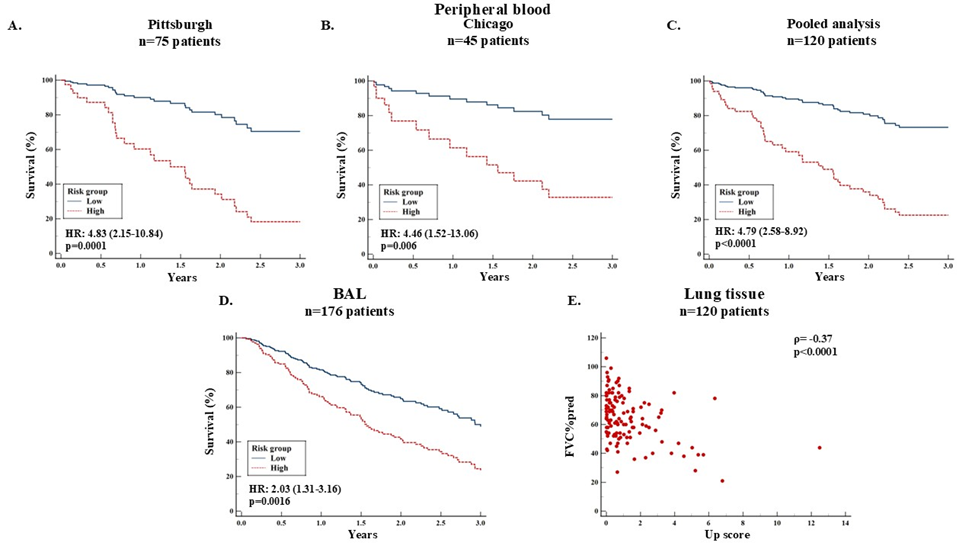
Prognostic accuracy of the 6-gene profile in different cohorts and sample types: A 6-gene profile (BPI, FCAR, HK2, IMPA2, S100A12 and SERINC2) was generated using LASSO in Pittsburgh cohort and was validated in other cohorts. The value>1.82 was identified the threshold for mortality, which discriminates patients into high and low risk. Patients in the high-risk group had increased mortality risk compared to the low-risk group in Pittsburgh (Panel A), Chicago (Panel B) and in the pooled analysis (Panel C). Based on the same threshold as in the peripheral blood, patients in the high-risk group of the BAL cohort had significantly worse survival compared to the low-risk patients (Panel D). Spearman's coefficient of rank correlation shows that up score of the 6-gene profile in lung tissue was negatively correlated with FVC%predicted (Panel E).
Desired Partnerships
- License
- Sponsored Research
- Co-Development